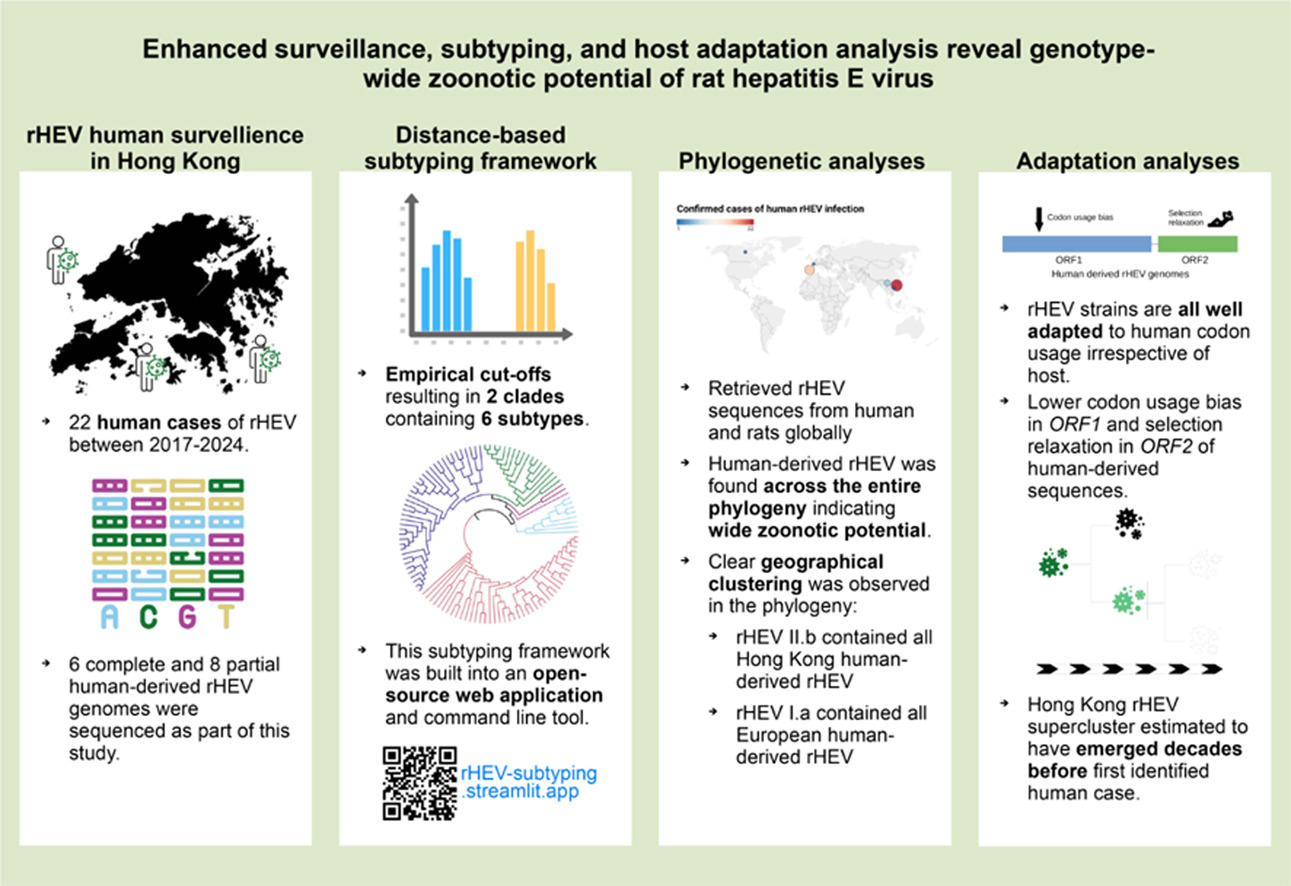
Rat hepatitis E virus (rHEV, Rocahepevirus ratti genotype 1) is a zoonotic pathogen that we have shown to be capable of infecting humans. A major hurdle in rHEV research is discrepancies between classification proposals. With GenBank deposits and new human-derived sequences from Hong Kong, a recent study led by Dr. Siddharth Sridhar proposes a nucleotide distance-based classification of an up-to-date rHEV genomic dataset, using maximum likelihood on top of usual substitution models to demonstrate the presence of six subtypes (a to f) and two clades (I or ade, II or bcf) in the phylogeny. An open-source web application for rHEV subtyping is available for public use at https://rhev-subtyping.streamlit.app/.
Clade I strains are more cosmopolitan and clade II strains appear localised to Southeast Asia. Even though Hong Kong has the most reported human cases and most of the strains form a supercluster in clade II subtype b, human-derived strains are found across both clades, in four out of six subtypes, indicating genotype-wide zoonotic potential.
Human-derived strains are then compared with animal-derived strains and subjected to codon usage bias (CUB) analysis. In the open reading frame 1 (ORF1) region, human-derived strains are found to be less biased in codon usage in the sense of having less preference in using certain codons (see section on ENC). Similarly, rHEV as a whole does not have a significantly higher bias to human codon usage than to other animal species (see section on CAI and n-CAI). These suggest rHEV, especially human-derived ones, are not specifically adapted to rodents and should be seen as a constant zoonotic threat to humans.
The team also used the RELAX model to analyse rHEV strains and concluded that the capsid-coding ORF2 region in human-derived strains is under weaker purifying selective constraints. It is more tolerant to the accumulation of nonsynonymous mutations, which may be a factor in its zoonotic ability. Observation on ORF3 having the highest pervasive positive selection is resonant of previous Paslahepevirus balayani HEV studies.
Finally, the researchers used a Bayesian approximation of the rHEV phylogeny in inferring the most recent common ancestor (MRCA). The MRCA of the Hong Kong supercluster is placed around the 1980-90s, decades before the first confirmed case of human rHEV infection. These strains have probably been circulating in rats and humans for several years before emerging to public health attention.
Read the full article: Journal of Hepatology. 2025 Jul 14:S0168-8278(25)02339-6. DOI: 10.1016/j.jhep.2025.07.004